Application of digital PCR in monitoring of non-invasive cancer treatment
Article by Dr. Le Thi Thanh Huong - Vinmec Stem Cell Research Institute and Gene Technology.
In this article, we will learn the application of digital PCR (Droplet Digital TM PCR, ddPCRTM) for liquid biopsy procedures in cancer diagnosis, monitoring and evaluation of treatment methods as well as development show recurrence after remission.
1. Summary
Liquid biopsy is a noninvasive method used to diagnose and monitor conditions, including cancer. Instead of biopsies of affected tissue, “cell-free DNA” (cfDNA), circulating tumor DNA (ctDNA) is collected; a subset of cfDNA and derived from within the tumour), or suspension cancer cells (also called circulating tumor cells, CTCs) from blood or other body fluids. Digital PCR (Droplet Digital TM PCR, ddPCRTM) is used for liquid biopsy procedures in cancer diagnosis, monitoring and evaluation of treatment modalities, and detecting recurrence after remission.
2. What is a liquid biopsy?
Liquid biopsies are noninvasive tests that detect pieces of DNA or cells in blood or other body fluids. The term "free DNA" (cfDNA) refers to fragments of DNA primarily derived from apoptotic and necrotic cells. Circulating cfDNAs can be used to diagnose a wide range of diseases including cancer, diabetes, and even myocardial infarction, as well as monitor transplanted organs in recipients. In the case of cancer, cfDNA is often referred to as circulating tumor DNA, or ctDNA.

cfDNA lưu hành trong máu
3. Application of liquid biopsy in diagnosis and monitoring of cancer treatment effectiveness
Today, cancer research and diagnosis is one of the important applications of liquid biopsy. This technical group helps to solve the following specific tasks.
Monitor and define treatment regimens – Liquid biopsies can help doctors monitor the effectiveness of a treatment regimen, optimize regimens, determine if changes in treatment regimens are harmful. benefit or not. Helps track changes in tumors – Many tumors change continuously over time; Monitoring these changes can help doctors recommend changes in treatment. Recurrence monitoring – Helps screen patients in remission to detect new tumor growth before physical signs become apparent, whether the tumor is present in the same site or has metastasized other mind. Regular screening of high-risk patients – For some people with a cancer genotype, a liquid biopsy can help detect cancer early before clinical symptoms appear or are detected by other methods. common photography techniques.
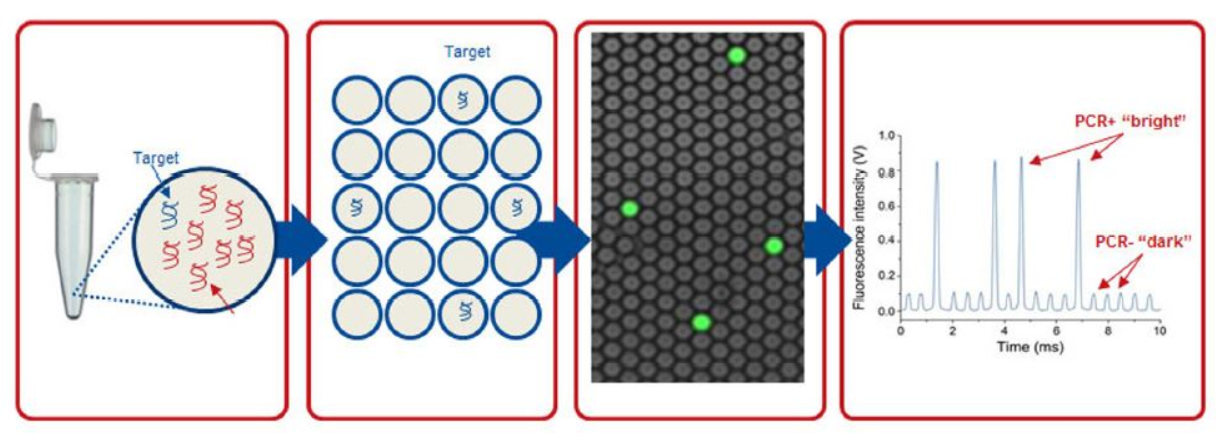
Công nghệ PCR (ddPCR)
The goal of liquid biopsy is to detect and quantify circulating mutated cfDNA, ctDNA and CTCs. However, these target sequences are usually present at low levels and are surrounded by other complex components such as white blood cells, normal DNA from aging and frail body cells, etc. In addition, Cyclic DNA is often highly fragmented, so the content of intact target sequences will not be high. These challenges call for more sensitive mutation screening techniques that can detect and quantify low concentrations of target sequences in a short time. Digital PCR or digital microdroplet PCR (Droplet DigitalTM PCR, ddPCRTM) is currently a suitable tool for liquid biopsies. This is a new technology that allows absolute quantification of target sequences with high sensitivity and accuracy.
4. Digital microdroplet PCR (ddPCR)
A digital microdroplet PCR procedure consists of 5 basic steps.
PCR reaction preparation: The preparation of a ddPCR reaction is similar to that of a conventional PCR reaction. The reaction also includes basic components such as Taq polymerase enzyme, primers, dNTP, magnesium, etc., and the addition of EvaGreen fluorescence or TaqMan fluorescent probe. DNA samples are added to these reactions. Just like when running PCR, in addition to reactions for the test sample, technicians often run positive control reactions (containing target DNA) and negative control reactions (containing no target DNA). Microdroplet generation: Using a special device called a Droplet Generator, a reaction is randomly divided into microdroplets with a volume of about 1 nL. Each of these microdroplets contains all the components of a Real-time PCR reaction. The DNA molecules will be randomly distributed into these microdroplets. Cloning of DNA contained in microdroplets: All microdroplets generated from an initial reaction are transferred to 1 well in a 96-well PCR plate and placed in a conventional PCR machine. When the heat cycle takes place, the DNA contained in each microdroplet is cloned as a conventional PCR reaction. The fluorescence signal produced by the probe or the fluorescence will be accumulated in each microdroplet, Read the signal in the microdroplet: After the PCR reaction is completed, the microdroplet will be analyzed by a fluorescence reader. optical. Each micro-droplet in each tube contained in step 3 is aspirated into the tube of the instrument and in turn passes through the fluorescence signal readers. The signal intensity of each fluorescent color present in each microdroplet will be recorded by the reader. Data analysis: The readings of all microdroplets generated from each initial reaction are transferred to the computerized analysis software. For each initial 20 μL reaction, the analyzed results included:
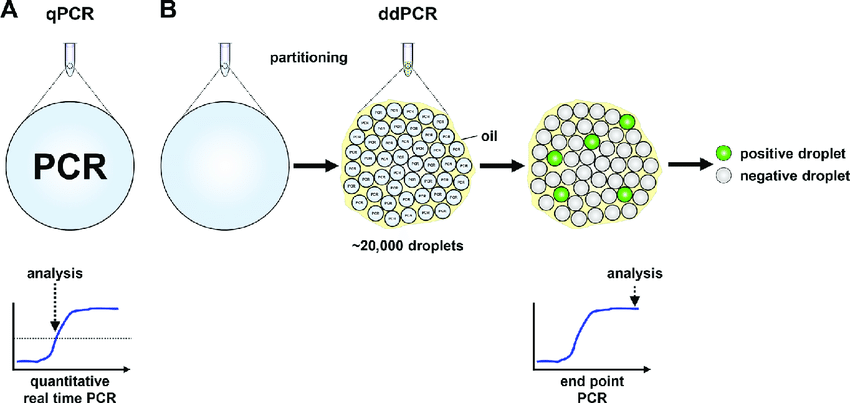
PCR vi giọt kỹ thuật số
Total number of droplets produced from the initial reaction, including positive and negative microdroplets. The total number of positive microdroplets (containing the target DNA and having a fluorescence signal intensity significantly higher than those in the negative control reaction). Total number of negative microdroplets (containing no target DNA and having the same fluorescence signal intensity as those in a negative control reaction) Based on the number of positive microdroplets and the total number of droplets produced from each reaction In the initial reaction, the software will apply the Poisson distribution model to calculate the target DNA concentration in the initial reaction volume.
5. Digital PCR application in cancer detection and monitoring
Today, there are many works in the world that have been applying digital PCR in detecting and monitoring many different types of cancer. Through the examples below, readers will visualize the applications of digital PCR as well as the excellent sensitivity of this technique.
Non-small cell lung cancer
Non-small cell lung cancer (NSCLC) is the most common form of lung cancer. NSCLC carries mutations that activate epidermal growth factor receptor (EGFR) and is responsive to tyrosine kinase inhibitors. There are many different EGFR-activating mutations, and after a period of treatment, the majority of NSCLCs become resistant to kinase inhibitors. This resistance is caused mainly by the secondary mutation EGFRT790M. Therefore, in addition to identifying EGFR mutations, doctors now think that it is necessary to check for the occurrence of secondary mutations during treatment. In their 2015 work, Zhu et al. applied digital PCR to analyze ctDNA and helped increase the clinical sensitivity for EGFR-activating mutations by 80%. In addition, digital PCR allows the detection of EGFRT790M secondary mutations at a low rate (Oxnard et al., 2014, Watanabe et al., 2015, Zhang et al. 2015) before the tumor re-grows. due to resistance to tyrosine kinase inhibitors.

Ung thư phổi không tế bào nhỏ (NSCLC)
Melanoma
Approximately 50% of melanoma skin cancers carry a mutation that converts valine to glutamic acid at codon 600 of the serine/threonine kinase B-Raf (a proto-oncogene). Patients carrying this mutation are treated with a BRAF inhibitor. The level of circulating cfDNA in blood containing this mutation is very low (<0.01% of cfDNA). However, in the past time, there has been a work to apply digital PCR to detect this type of BRAFV600E mutation and achieve a sensitivity of 0.005%. This work also applied digital PCR to demonstrate the correlation between the percentage levels of BRAFV600E mutations in tumors and in cfDNA (Sanmamed et al., 2015). In addition, in patients treated with BRAF inhibitors, a decrease in BRAFV600E ctDNA would correlate with tumor destruction, and conversely an increase in BRAFV600E ctDNA would be an indicator of resistant to treatment. The study by Sanmamed et al. has demonstrated that it is possible to apply digital PCR to identify patients who are likely to respond to BRAF inhibitor therapy as well as monitor the treatment process. theirs afterwards.
Bệnh nhân bị ung thư da
Breast cancer
In breast cancer , because the recurrence rate after surgery is significant, doctors may prescribe additional treatment with radiation therapy and/or chemotherapy to destroy the tumor cells you are left over. Beaver et al applied digital PCR to analyze ctDNA samples before and after breast cancer surgery. The research team detected ctDNA in several postoperative samples (Beaver et al., 2014). This suggests that micro-metastasis is still present in these samples and that we can apply digital PCR to detect residual solid tumors (Siravegna and Bardelli, 2014). From this, we can see the feasibility of applying digital PCR to detect low levels of ctDNA in order to predict the risk of recurrence after breast cancer surgery and to inform decisions about the type of after-care. suitable surgery.
Recommended video:
References Beaver JA et al. (2014). Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin Cancer Res 20, 2643–2650. PMID: 24504125 Oxnard GR et al. (2014). Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 20, 1698–1705. PMID: 24429876 Sanmamed MF et al. (2015). Quantitative cell-free circulating BRAFV600E mutation analysis by use of droplet digital PCR in the follow-up of patients with melanoma being treated with BRAF inhibitors. Clin Chem 61, 297–304. PMID: 25411185 Siravegna G and Bardelli A (2014). Minimal residual disease in breast cancer: in blood veritas. Clin Cancer Res 20, 2505–2507. PMID: 24658155 Watanabe M et al. (2015). Ultra-sensitive detection of the pretreatment EGFR T790M mutation in non-small cell lung cancer patients with an EGFR-activating mutation using droplet digital PCR. Clin Cancer Res epub ahead of print. PMID: 25882755 Zhang B et al. (2015). Comparison of droplet digital PCR and conventional PCR for measuring EGFR gene mutations. Exp Ther Med 9, 1383–1388. PMID: 25780439 Zhu G et al. (2015). Highly sensitive Droplet Digital PCR method for detection of EGFR-activating mutations in plasma cell-free DNA from patients with advanced non-small cell lung cancer. J Mol Diagn 17, 265–272. PMID: 25769900 MORE:
Learn about cancer diagnostic ctDNA liquid biopsy Cervical intraepithelial neoplasia (CIN): What you need to know How do cancer cells form, grow and spread any?
Bài viết này được viết cho người đọc tại Sài Gòn, Hà Nội, Hồ Chí Minh, Phú Quốc, Nha Trang, Hạ Long, Hải Phòng, Đà Nẵng.




